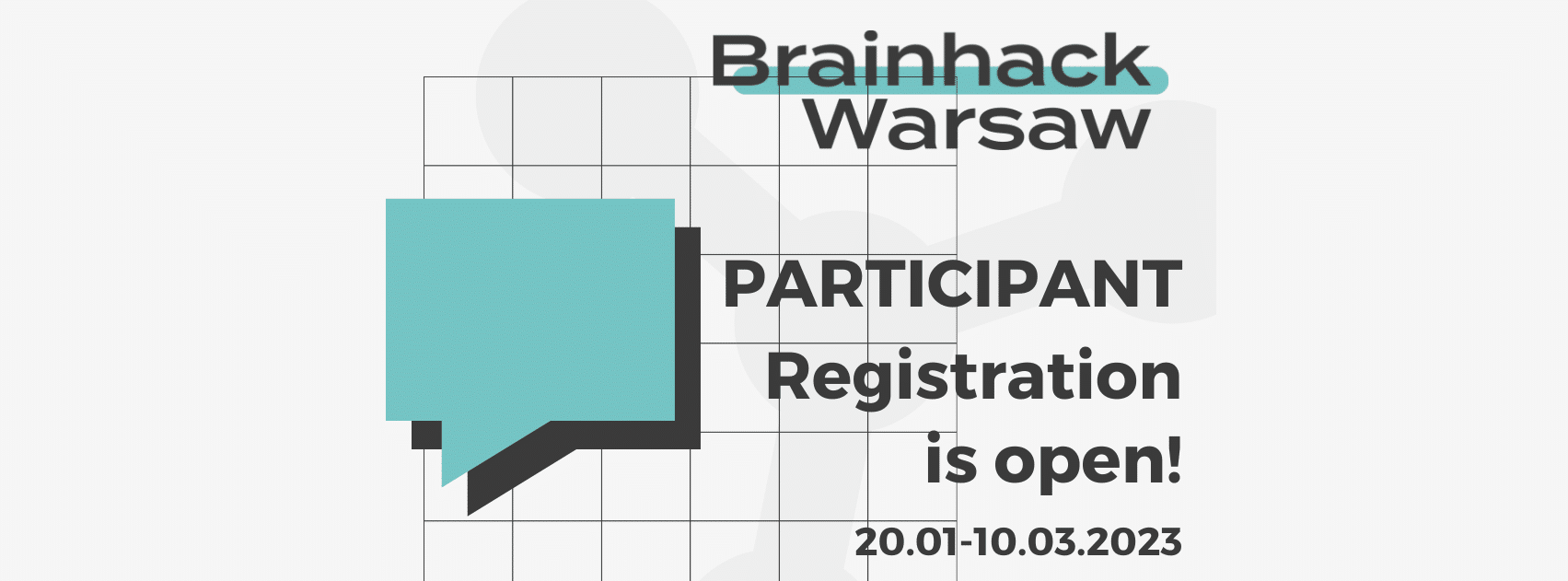
About Brainhack Warsaw 2023
On the third weekend of March 2023, the fifth edition of Brainhack Warsaw will take place. During this three-
day event dedicated to students and PhD students, we will work in teams on neuroscience-related projects.
This year’s edition is organized by the Neuroinformatics Student Club operating at Physics Faculty, University of Warsaw.
The aim of the event is to meet new, enthusiastic researchers, make new friendships in academia, learn, share
the knowledge on data mining, machine learning and brain research, but also promote open science in the
spirit of the whole Brainhack community (Craddock et al., 2016). Attendees of various backgrounds are welcome to join!
By submitting your own, genuine research project, you can gain a priceless leadership experience as you will
manage a group of researchers at our three-day event. Go ahead, let your creativity bloom and share your idea with us!
Deadline for project proposals: 6.01.2023
Announcement of projects: 15.01.2023
Participant registration starts: 16.01.2023
Deadline for participant registration: 3.03.2023
Please send all the related questions at the mailing address
One on the main establishments for Brainhack Warsaw is to provide a safe and comfortable experience for
everyone, regardless of their gender, gender identity and expression, sexual orientation, disability, physical appearance,
body size, race, age or religion. Please make sure to familirize with a Code of Conduct of the Brainhack Global.
If you vitness any form of harassment at our event, please inform us at our (e-mail adress)
so we can react and prevent any future events like that.
Dodaj tu swój tekst nagłówka
Past Editions
This is the fifth time that Brainhack Warsaw is happening. We are proud that out initiative has gone so far and happy to share with you what have happened before. You can read about all the projects conducted before, engage with our previous guests and also get a glimpse of what you might be expecting this year.
Remember that reading all about what we experienced before might be a great start on your journey with Neuroinformatics, Machine Learning and Brain Sciences. Reading about previous editions you can also be inspired to prepare Brainhack Warsaw 2023 project idea.
We believe that the drive for exploration, curiosity and reaching out is what makes our community a great place for everyone. See what it was and is like to be a part of this initiative and do not hesitate to contact us in case of any further questions conserning previous editions of our hackathon conference.
Click on our brain and find out more about Brainhack Warsaw!
Speakers

Deep Learning Tools for the Analysis of Movement, Identity & Behavior
Abstract: Quantifying behavior is crucial for many applications in biology and beyond. Videography
provides excellent methods for the observation and recording of animal behavior in diverse
settings, yet extracting particular aspects of a behavior for further analysis can be highly time
consuming and computationally challenging. I will discuss the latest developments for
DeepLabCut, an efficient method for markerless pose estimation based on transfer learning
with deep neural networks that achieves excellent results with minimal training data (Mathis et
al., Nature Neuroscience 2018; Nath et al., Nature Protocols 2019). For multiple animals, I will
discuss animal-agnostic assembly and tracking methods as well as the ability to predict an
animal’s identity from the same backbone to assist tracking and perform Re-Identification
(Lauer et al. Nature Methods, 2022). I will illustrate the versatility of these tools for multiple
species across a broad collection of behaviors from egg-laying flies, via centipedes to 3D
pose estimation on hunting cheetahs.
Bio: Alexander studied pure mathematics with a minor in logic and theory of science at the Ludwig Maximilians University in Munich. For his PhD also at LMU, he worked on optimal coding approaches to elucidate the properties of grid cells. As a postdoctoral fellow with Prof. Venkatesh N. Murthy at Harvard University and Prof. Matthias Bethge at Tuebingen AI, he decided to study olfactory behaviors such as odor-guided navigation, social behaviors and the cocktail party problem in mice. During this time, he increasingly got interested sensorimotor behaviors beyond olfaction and started working on proprioception, motor adaption, as well as computer vision tools for measuring animal behavior.

Michal Niezgoda
CEO & Co-Founder at Robotec.ai
Projects
Project 1: Brain Tumor Subtyping
Authors: Jędrzej Kubica 1
- University of Warsaw
Abstract: Brain tumors can be classified into different subtypes to recommend a patient-specific treatment. Characterization of patients into groups based on molecular features can help clinicians to choose proper medications for each individual and to improve the outcome of the treatment. Various classification tools for tumors of the central nervous system have been developed. For instance, MethPed [1] is an open-access classifier which uses DNA methylation as an epigenetic marker for subtype classification. The goal of the project is to develop of a data analysis workflow to analyze publicly-available epigenetic data of brain tumor patients from databases, such as The Cancer Genome Atlas. Future work will include subtype-specific drug recommendations. Further research can also be extended from brain tumors into tumors of the nervous system.
[1] Danielsson, A., Nemes, S., Tisell, M. et al. MethPed: a DNA methylation classifier tool for the identification of pediatric brain tumor subtypes. Clin Epigenet 7, 62 (2015). https://doi.org/10.1186/s13148-015-0103-3
A list of 1-5 key papers/materials summarising the subject:
- https://doi.org/10.1073/pnas.220201511
- https://doi.org/10.1093/annonc/mdv024
- https://doi.org/10.1186/s13148-015-0103-3
A list of requirements for taking part in the project:
- basics of R would be an advantage
- basics of Python would be an advantage
A maximal number of participants: 15
Skills and competences you can learn during the project:
- How to use epigenetic data in cancer research
- How to retrieve cancer data
- How to create bioinformatic data analysis workflows
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 2: Automatic artifact detection in EEG using Telepathy
Authors: Marcin Koculak 1
- Consciousness Lab, Institute of Psychology, Jagiellonian University
Abstract: This will be a follow-up to a project from Brainhack Warsaw 2022, where we built a library for EEG analysis from scratch. That library since then was named „Telepathy” and is being actively developed by the author of this project. In this edition I would like to focus on implementing methods for automatic detection of artefactual signal in EEG recording, along with methods to deal with them. This includes correcting hardware related issues, rejecting signals coming from non-brain sources, and methods for dimensionality reduction, all of which help to isolate the signal of interest from EEG. Participants will have a chance to better understand sources of noise in EEG data, see what analysis software does under the hood to deal with them, and attempt to implement them in new and dynamically evolving language for scientific computing. If possible, I will try to organise a small demonstration how EEG signal is collected and from where the artefacts might come from.
A list of 1-5 key papers/materials summarising the subject:
- Telepathy website: https://github.com/Telepathy-Software/Telepathy.jl
- Online materials from other EEG software, e.g. MNE-Python: https://mne.tools/stable/auto_tutorials/preprocessing/10_preprocessing_overview.html
A list of requirements for taking part in the project:
- Basic understanding of data analysis, signal processing, and brain physiology will make the project much more approachable for users, but everyone is welcomed
- Prior experience with programming (especially in Julia, Python, or Matlab) will also help
A maximal number of participants: 15
Skills and competences you can learn during the project:
- Better understanding of EEG data collection and processing
- Sources of artefacts and methods to deal with them
- Basics of programming in Julia
- Collaboration on a software project with version control (git).
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 3: Application of an amygdala parcellation pipeline based on Recurrence Quantification Analysis to resting-state fMRI data acquired in a 7T MRI scanner
Author: Sylwia Adamus1
- Medical University of Warsaw, University of Warsaw Faculty of Physics
Abstract: According to animal studies the amygdala consists of several groups of nuclei, which play different roles in emotion-related processes. It has also been shown that this brain structure is important for the development of many psychiatric conditions, such as depression, addictions, and autism spectrum disorder. Up to this day, a number of approaches to the topic of amygdala parcellation have been suggested. One of them, which was recently published, is a pipeline using Recurrence Quantification Analysis (RQA). It enables the division of the human amygdala into two functionally different parts based on brain signal dynamics [1]. The aim of this project is to further develop this pipeline and check whether with its help it is possible to divide the amygdala into more than two subdivisions. To achieve this the pipeline will be applied to resting-state fMRI data acquired in a 7T MRI scanner from a dataset consisting of 184 healthy subjects from the Human Connectome Project. An exploratory approach will be applied using several variations of RQA parameters and clustering algorithms. The pipeline was developed on resting-state fMRI data acquired in a 3T MRI scanner. It has been speculated, that its application to data from a 7T MRI scanner could enable obtaining more detailed parcellations. Therefore the main hypothesis behind this project is that by using this pipeline it will be possible to achieve a parcellation with at least three functionally different subdivisions. It could have the potential to serve as a mask in further studies of human amygdala functional connectivity. All participants will have the opportunity to go through the whole pipeline to fully explore its possibilities. A device, which supports Anaconda Navigator is necessary to take part in the project. No advanced neuroscientific knowledge is required and everyone, who knows some basics of Python programming, is invited to cooperate. References: [1] Bielski K. et al., (2021) NeuroImage 227(117644)
A list of 1-5 key papers/materials summarising the subject:
A list of requirements for taking part in the project:
- Ability to programme in Python is an important requirement for this project (solid basics will be enough).
- Knowledge of Matlab will be appreciated, however it is not a must.
- English level, which enables effective communication between team members and reading scientific manuscripts, will be satisfactory.
- Everyone, who wants to participate, will need to have a device which supports Anaconda Navigator.
A maximal number of participants: 4
Skills and competences you can learn during the project:
- Project participants can learn the basics of Recurrence Quantification Analysis and how it can be applied to neuroimaging data.
- They will also have the opportunity to directly cooperate with the pipeline’s co-author and gain knowledge on using clustering algorithms and computing internal validation measures.
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 4: Pose estimation based long term behavior assesement of animals in semi-naturalistic condtions
Author: Konrad Danielewski1, Marcin Lipiec, Ewelina Knapska, Alexander Mathis
- Nencki Institute of Experimental Biology
Abstract: Using Eco-HAB system of four cages equiped with RFID antennas (RFID tagged group of 12 mice) and DeepLabCut, SOTA pose estimation framework we want to create a framework for behavior analysis of those animals. A set of 9 bodyparts is tracked on each animal (pose estimation model is ready and working well). Our test recording is 3 hours long but the goal is for the system to perform non-stop for a week. A synchronization script that will work based on visual information is needed, as there is no TTL signal from the antennas – camera timestamps are available and between frame times are very precise (at the scale of 10s of microseconds – FLIR BFS camera is used). We aim to develop an algorithm that will filter identity based on antenna readout and correct any potential switches. With identity corrected DLC2Kinematics and DLC2Action with some additional functions can be used to assess animal behavior for single animals and pairwise social interations. The goal would be to combine all of this into a user friendly framework with a set of high-level functions and thorough documentation that can be open-sourced and used by anyone interested in highly detailed long-term, homecage monitoring and behavior assesement.
A list of 1-5 key papers/materials summarising the subject:
- Lauer, J., Zhou, M., Ye, S., Menegas, W., Schneider, S., Nath, T., … & Mathis, A. (2022). Multi-animal pose estimation, identification and tracking with DeepLabCut. Nature Methods, 19(4), 496-504
- Mathis, M. W., & Mathis, A. (2020). Deep learning tools for the measurement of animal behavior in neuroscience. Current opinion in neurobiology, 60, 1-11
- Puścian, A., Łęski, S., Kasprowicz, G., Winiarski, M., Borowska, J., Nikolaev, T., … & Knapska, E. (2016). Eco-HAB as a fully automated and ecologically relevant assessment of social impairments in mouse models of autism. Elife, 5, e19532.
A list of requirements for taking part in the project:
- Participants should be proficient in English and data analysis
A maximal number of participants: 4
Skills and competences you can learn during the project:
- Participants can learn how to approach a project that is aimed to be open-source and can be used by people that have minimal coding experience. It’s a great way to start working with complex pose estimation data and new behavior segmentation frameworks.
Is there a plan for extending this work to a paper in case the results are promising? Not sure
Project 5: A paradigm shift in experiment design leading to large scale EEG data acquisition for visual attention
Author: Shaurjya Mandal1
- Carnegie Mellon University
Abstract: Visual attention often refers to the cognitive processes that allow the selective processing of visual information that we are exposed to in our daily lives. Gaining an understanding of visual attention can be crucial to a number of applications like the study of human-computer interaction and analysis and improvement of advertisements. EEG along with eye-tracking has been popularly used to study visual attention in subjects. In deep learning applications, to model needs to be trained on a significantly large dataset. Although the results obtained with deep learning algorithms tend to be highly accurate, it is always hard to acquire such a dataset. The apparatus required to track the eye movements and comment on visual attention from the gaze of the subject are not portable. Thus, to sync the EEG data with eye tracking/ visual attention data outside the laboratory setup, we would need an additional approach. To study the same, we ask two important questions:
1. To what extent can multi-channel EEG data provide inference regarding eye movement and eye tracking? And are there any specific experiments that help us to observe the behaviour better?
2. Is there a way to find an alternative to proper eye-tracking that can be crowdsourced? To answer the above questions, we would start with analysing 3 distinct publicly available datasets.
EEGEyeNet dataset comprises of EEG and eye-tracking data from 350 participants, 190 female and 160 male, between the ages of 18 and 80 years. This dataset provides enough data to correlate the changes in EEG based on the movement of the eyes. The eye-tracking experiments with synchronized EEG have been divided into 3 main parts: Pro and antisaccade, large grid, and visual symbol search. Knowing the protocols of the experiment while analysing the data would allow gain a better understanding of eye-movement with the EEG data. In the previous literature, eye-tracking for visual attention is linked to mouse-tracking. But to allow this, specific protocols have to be adopted that causes takes care of localised visual exposure with minimal distraction. To validate our performance across eye-tracking and mouse-tracking scenarios, we will make use of the OSIE dataset. OSIE dataset consists of 700 natural images along with publicly available mouse tracking and eye tracking data. To train the deep learning model that will allow us to generate labels for visual attention, we have made use of the SALICON dataset. The dataset consists of 10,000 MS COCO training images. By the end of the project, we hope to:
1. Analyse the pretrained models for visual attention with eye tracking and train our own models with the mouse-tracking data and compare the models.
2. Through effective data analysis, reduce the number of channels while preserving the variance of EEG data during eye-tracking experiments.
3. Develop our own custom software which can be used as a convenient means to collect and sync EEG data to visual attention outside laboratory settings.
A list of 1-5 key papers/materials summarising the subject:
1. Kastrati, Ard, et al. ”EEGEyeNet: a Simultaneous Electroencephalography and Eye-tracking Dataset and Benchmark for Eye Movement Prediction.” arXiv preprint arXiv:2111.05100 (2021).
2. Xiang, Brian, and Abdelrahman Abdelmonsef. ”Too Fine or Too Coarse? The Goldilocks Composition of Data Complexity for Robust Left-Right Eye-Tracking Classifiers.” arXiv preprint arXiv:2209.03761 (2022).
3. Xu, Juan, et al. ”Predicting human gaze beyond pixels.” Journal of vision 14.1 (2014): 28-28.
4. Jiang, Ming, et al. ”Salicon: Saliency in context.” Proceedings of the IEEE conference on computer vision and pattern recognition. 2015.
5. Lou, Jianxun, et al. ”TranSalNet: Towards perceptually relevant visual saliency prediction.” Neurocomputing 494 (2022): 455-467.
A list of requirements for taking part in the project:
- All the participants should be able to communicate in English (basic communication is fine)
- ideally the participants could be doing their undergraduate or graduate studies or be part of an industry but having a background in deep learning or signal processing (especially in biosignals)
A maximal number of participants: 8
Skills and competences you can learn during the project:
- The participants shall gain exposure regarding the applications of machine learning and deep learning in signal and image processing. They would learn about the decision metrics that helps to validate the performance of a model. The project would extensively discuss the variations of experiment design adopted to reach a final goal. By the end, the participants would be able to design their own data acquisition experiments.
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 6: Robust Latent Space Exploration of EEG Signals with Distributed Representations
Author: Adam Sobieszek1
- University of Warsaw
Abstract: It is said that the advantage of methods, such as Variational Autoencoders (VAEs) and Generative Adversarial Networks (GANs), is their learning of latent space representations of the target domain. E.g. training a GAN that generates EEG signals gives us a latent space representation of the type of EEG signals that were in the training set. These representations could moreover be made „disentangled”, which uncovers independent dimensions that are features that best describe the target domain. Such methods could give us the ability to describe and better understand the type of information contained in EEG signals. However, for such methods to be widely accepted as scientific tools, we need to overcome the issue that each time we train a neural network, the learnt latent space seems on the surface to be completely different. Recently we investigated the ability of GANs to learn disentangled representations of EEG signals. With our network, we’ve obtained multiple latent space representations of EEG signals. The goal of this project is to investigate whether such representations hold the same kind of information, possibly showing the robust nature of neural network representation learning, as well as investigate how such multiple (distributed) representations may be used together. We will implement down-stream tasks, that latent representations are useful for (such as classification, and explainable feature visualisation) and compare how their performance differs between different representations. We will investigate whether combining representations may result in an increase in accuracy. We will test methods for learning probabilistic maps between latent spaces, which could prove useful for a wider array of machine learning applications. Finally, we will attempt to find in these distributed representations the common factors, that are robustly able to describe EEG signals. If time allows we will also attempt to train disentangled variational autoencoders (called beta-VAEs) for generating EEG signals and investigate whether the factors discovered by this method are similar to those found with our GAN-based method.
A list of 1-5 key papers/materials summarising the subject:
- EEG-GAN, that our network is based on: Hartmann et al. 2018.
- Disentanglement in GANs was introduced in: Karras et al. 2020.
- Disentanglement in VAEs: Burgess et al., 2018
A list of requirements for taking part in the project:
- For coding people: machine learning in python (sklearn or pytorch)
- For non-coding people: EEG analysis
A maximal number of participants: 10
Skills and competences you can learn during the project:
- Learn some theory behind representation learning, pratice implementing down-stream tasks that use latent space representations, create together new tools for combining latent space representations
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 7 SANO: Comparison brain networks from EEG and fNIRS
Authors: Rosmary Blanco, Cemal Koba, Alessandro Crimi
- Sano center for computational medicine
Abstract: The integrated analysis of functional near infrared spectroscopy (fNIRS) and electroencephalography (EEG) provides complementary information about electrical and hemodynamic activity of the brain. Evidence supports the mechanism of the neurovascular coupling mediating their brain processing. However, it is not well understood how the specific empirical patterns of neuronal activity measured by these techniques underlies brain function and networks. Here we have compared the functional networks of the whole brain between synchronous EEG and fNIRS connectomes across frequency bands, using source space analysis. We have shown that both EEG and fNIRS networks have a small-world topology. We have also observed an increased interhemispheric connectivity for HbO compared to EEG and HbR, with no differences across the frequency bands. Our results demonstrate that some topological characteristics of the networks captured by fNIRS and EEG are different. Once we understand what their differences and similarities mean in order to interpret them correctly, their complementarity could be useful for clinical application.
A list of 1-5 key papers/materials summarising the subject:
- Chiarelli et al., “Simultaneous functional near-infrared spectroscopy and electroencephalography for monitoring of human brain activity and oxygenation: a review,” Neurophotonics, vol. 4, no. 4, pp. 041411, 2017
2.Yuxuan Chen et al., “Amplitude of fnirs resting-state global signal is related to eeg vigilance measures: A simultaneousfnirs and eeg study,” Frontiers in Neuroscience, vol. 14, 2020.
- Jaeyoung Shin et al., “Open access dataset for EEG + NIRS single-trial classification,” IEEE TNSRE, vol. 25, no. 10, pp. 1735–1745, 2016
- M. Elsheikh, Relating Global and Local Connectome Changes to Dementia and Targeted Gene Expression in Alzheimer’s Disease, Frontiers in human neuroscience, 2021
A list of requirements for taking part in the project:
- Basic understanding of data analysis, signal processing, and brain physiology
- Prior experience with programming in Python/Matlab
- Knowledge of complex networks is an asset
A maximal number of participants: 10
Skills and competences you can learn during the project:
- Better understanding of computational neuroscience
- A new portable modality complementary to EEG
Is there a plan for extending this work to a paper in case the results are promising? Yes
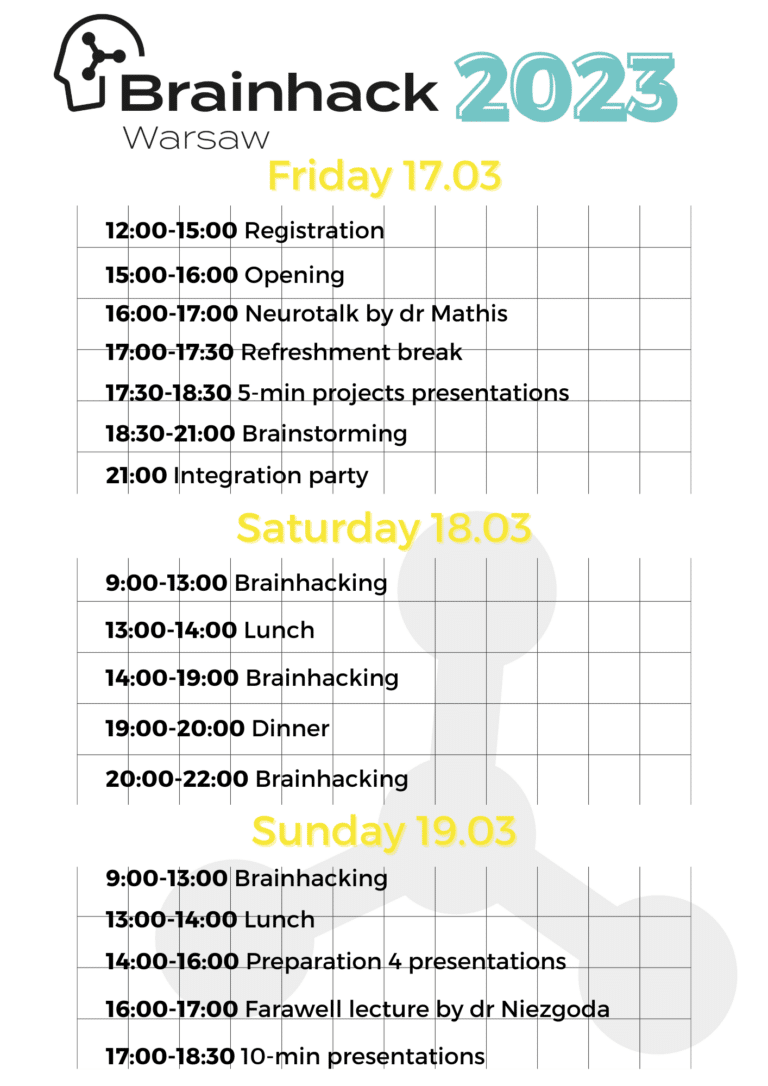
Schedule
Participant Registration
The registration for Brainhack Warsaw 2023 is open.
Register here —> https://forms.gle/gGPaXVXMyYWwLzCT9
Registration took place in two rounds and there was a small registration fee (to cover the catering during the event) for the project participants
EARLY -> 200 PLN (around 43 EUR) from TBA to 17th February
REGULAR -> 225 PLN (around 48 EUR) from 18th February to 3rd March
Please keep in mind that all of the payments should be made in PLN (polish złoty).
Partners & sponsors
Committee

Julia Jakubowska
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Łukasz Niedźwiedzki
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Zofia Mizgalewicz
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Ada Kocholewska
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Weronika Plichta
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Aleksandra Bartnik
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Przemysław Ziółkowski
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Julia Jurkowska
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Marcin Syc
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Agata Kulesza
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland
Sylwia Adamus
Neuroinformatics,
Medical University of Warsaw, Faculty of Physics, University of Warsaw,
Warsaw, Poland

Krzysztof Wróblewski
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Maria Waligórska
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Szymon BociaN
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Emilia Kaczmarczyk
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland
Advisory board:

Jarosław Żygierewicz, PhD
Neuroinformatics,
Biomedical Physics Division,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

jacek Cyranka, PhD
Assistant professor in Computer Science,
Institute of Informatics,
Faculty of Mathematics, Informatics, and Mechanics, University of Warsaw,
Warsaw, Poland
Graphics
Piotr Gadoś
Venue
University of Warsaw, Faculty of Physics, Pasteura 5, 02-093 Warsaw, Poland.