About Brainhack Warsaw 2024
On the 15th to the 17th of March 2024, the sixth edition of Brainhack Warsaw will take place. During this three-
day event dedicated to students and PhD students, we will work in teams on neuroscience-related projects.
This year’s edition is organized by the Neuroinformatics Student Club operating at Physics Faculty, University of Warsaw.
The aim of the event is to meet new, enthusiastic researchers, make new friendships in academia, learn, share
the knowledge on data mining, machine learning and brain research, but also promote open science in the
spirit of the whole Brainhack community (Craddock et al., 2016). Attendees of various backgrounds are welcome to join!
By submitting your own, genuine research project, you can gain a priceless leadership experience as you will
manage a group of researchers at our three-day event. Go ahead, let your creativity bloom and share your idea with us!
Deadline for project proposals: 14.01.2024
Announcement of projects: 26.01.2024
Participant registration starts: 26.01.2024
Deadline for participant registration: 29.02.2024
Please send all the related questions at the mailing address.
One on the main establishments for Brainhack Warsaw is to provide a safe and comfortable experience for
everyone, regardless of their gender, gender identity and expression, sexual orientation, disability, physical appearance,
body size, race, age or religion.
If you witness any form of harassment at our event, please inform us at our (e-mail adress)
so we can react and prevent any future events like that.
Past Editions
This is the sixth time that Brainhack Warsaw is happening. We are proud that out initiative has gone so far and happy to share with you what have happened before. You can read about all the projects conducted before, engage with our previous guests and also get a glimpse of what you might be expecting this year.
Remember that reading all about what we experienced before might be a great start on your journey with Neuroinformatics, Machine Learning and Brain Sciences. Reading about previous editions you can also be inspired to prepare Brainhack Warsaw 2024 project idea.
We believe that the drive for exploration, curiosity and reaching out is what makes our community a great place for everyone. See what it was and is like to be a part of this initiative and do not hesitate to contact us in case of any further questions conserning previous editions of our hackathon conference.
Click on our brain and find out more about Brainhack Warsaw!
Projects
Registration deadline: 29.02.2024!
Project 1: Combining Generative Autoencoder and Complex-Valued Neural Network architectures for EEG signal modeling
Leader: Adam Sobieszek 1
- University of Warsaw
Abstract:The project aims to solve problems with EEG signal analysis with Variational Autoencoders (VAEs) by combining them with the lesser-known architecture of Complex-Valued Neural Networks (CVNNs). Our aim is to enhance the signal generation and representation capabilities of VAEs for EEG signals, developing a better architecture for modelling signals in the frequency domain, which is represented with complex numbers. VAEs are deep learning models that learn to encode and decode data, creating a latent space representation. This latent space is a compressed knowledge representation of the training signal data, which VAEs learn to reconstruct. In the context of EEG signals, VAEs are useful for signal generation (e.g. for missing data imputation) and their learnt representations can be used for clasification and prediction, as well as for uncovering insights about the training EEG data. Training VAEs on EEG data in the time domain has proven ineffective. Conversely, representing EEG signals in the frequency domain via their Fourier Transform is challenging due to these representations being complex numbers, that are not well transformed with real-valued neural network layers. CVNNs integrate complex numbers directly into the network architecture, using complex numbers in the trainable layer weights. In our project we will program custom CVNN layers that we can use to manipulate the complex-valued Fourier spectra. By combining VAEs with CVNNs, we can learn a complex-valued latent space representation of EEG signals, which can be interpreted in terms of the magnitude and phase of discovered signal components. We will train such networks on EEG data from psychological studies and analyze the learnt representation in a supervised and unsupervised setting. If successful, we expect to continue working on the project after the Brainhack in order to present the results in a scientific publication.
A list of 1-5 key papers/materials summarising the subject:
- https://www.youtube.com/watch?v=HxQ94L8n0vUhttps://doi.org/10.1073/pnas.220201511
- https://arxiv.org/pdf/2204.02195.pdf
- https://arxiv.org/pdf/2306.09827.pdf
A list of requirements for taking part in the project:
- knowledge of python and at least one of the following: familiarity with the pytorch package OR
- EEG signal processing in python OR
- understanding of mathematics of complex numbers OR
- fourier signal analysis
A maximal number of participants: 12
Skills and competences you can learn during the project:
- Theoretical understanding and practical experience with two cutting edge neural network architectures, potential for further collaboration and publication
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 2: Subtyping and grading of gliomas using artificial intelligence
Leader: Paulina Domek 1
- SWPS University
Abstract: Advances in brain tumor research show promise to improve diagnosis and treatment by identification of epigenetic molecular targets responsible for the disease (Skouras et al., 2023). It has been shown that gene expression is controlled by epigenetic modifications of DNA, such as histone methylation and acetylation (Gibney and Nolan, 2010), therefore, it is crucial to accurately identify molecular targets responsible for the occurrence of the disease. Recently, various approaches have been taken to combine transcriptomics (gene expression data analysis) and epigenetics to design novel drugs to target malfunctioning proteins, specific for each subtype and grade of brain tumors. The project was initiated at Brainhack 2023. In the project, we successfully developed a data analysis workflow to analyze publicly-available epigenetic data of brain tumor patients from various databases, such as The Cancer Genome Atlas and Gene Expression Omnibus. We validated an open-source software MethPed (Ahamed et al., 2016) as a potentially useful tool for clinical application of pediatric brain tumor subtyping. We found out that MethPed could serve as a confirmation test for the primary diagnosis, however some results might require additional confirmation. To emphasize the importance of code and data reproducibility, we shared our results in publicly available GitHub repository: https://github.com/jjjk123/Brain_tumor_subtyping. At Brainhack 2024, we plan to extend the workflow to include a machine learning software for subtyping and grading of gliomas using transcriptomic data (Munquad et al., 2022). The results will be analyzed alongside the results from MethPed, and are expected to provide a deeper insight into the mechanisms of brain tumor subtypes.
A list of 1-5 key papers/materials summarising the subject:
- Ahamed, M. T., Danielsson, A., Nemes, S., & Carén, H. (2016). MethPed: An R package for the identification of pediatric brain tumor subtypes. BMC Bioinformatics, 17(1), 262.
- Clarke, J., Penas, C., Pastori, C., Komotar, R. J., Bregy, A., Shah, A. H., Wahlestedt, C., & Ayad, N. G. (2013). Epigenetic pathways and glioblastoma treatment. Epigenetics, 8(8), 785–795.
- Gibney, E. R., & Nolan, C. M. (2010). Epigenetics and gene expression. Heredity, 105(1), 4–13.
- Munquad, S., Si, T., Mallik, S., Li, A., & Das, A. B. (2022). Subtyping and grading of lower-grade gliomas using integrated feature selection and support vector machine. Briefings in Functional Genomics, 21(5), 408–421.
- Skouras, P., Markouli, M., Strepkos, D., & Piperi, C. (2023). Advances on Epigenetic Drugs for Pediatric Brain Tumors. Current Neuropharmacology, 21(7), 1519–1535.
A list of requirements for taking part in the project:
- Biological or medical background would be an advantage
- Familiarity with Python or R would be an advantage
- Familiarity with GitHub would be an advantage
A maximal number of participants: No limit
Skills and competences you can learn during the project:
- Integration of transcriptomic and epigenetic data in cancer research
- How to retrieve and use cancer data
- How to build and use bioinformatic data analysis workflows
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 3: Visualization of Dyslexic Reading using Large Language Model
Leader: Karolina Źróbek1
- Akademia Górniczo-Hutnicza w Krakowie
Abstract: Large language models (LLMs) open up new possibilities, allowing us to explore and understand various perspectives of human experiences. The primary aim of this project is to create a visualization of dyslexic reading patterns predicted by a custom ChatGPT (Generative Pre-trained Transformer). By training a specialized language model on articles related to reading and dyslexia, we intend to develop a tool that can generate insightful analyses of dyslexic reading behaviors, including saccadic movements, fixation points, and the duration of fixations. Furthermore, the custom model will provide qualitative information on how individuals with dyslexia perceive text while reading. The information obtained from the custom language model can be used to create visualizations, such as videos or images, representing dyslexic reading patterns. Apart from enabling the viewer to empathize with dyslexic readers, we see a possible development of the visualization in such a way that provides dyslexic readers with a tool for enhanced comprehension of any given text. This project proposal is rooted in the exploration of eye movement patterns observed in individuals with dyslexia, coupled with qualitative research focusing on the subjective experience of reading. By delving into the intricate dynamics of eye movements among individuals facing dyslexia, we aim to unravel deeper insights into how this neurological condition shapes the act of reading. To achieve the stated objective, we propose the following implementation plan: Custom GPT Training: Training a custom Language Model (LM) using a diverse dataset of articles specifically focused on reading and dyslexia. The model would be used as a meaning and parts of speech identification tool, allowing for a comprehensive understanding of textual content. Text Analysis: The model would later be used to generate detailed text analyses including information related to saccadic movements, fixation points, and the length of fixations during dyslexic reading. The model would also provide qualitative insights into how dyslexic individuals perceive and interpret text while reading, capturing subjective experiences, such as struggles with specific words or patterns. Visualization Creation: The information obtained from the language model would be utilized to create visualizations representing dyslexic reading patterns. Prompt-to-image generative models could be used to enhance the visual representation of the data.
A list of 1-5 key papers/materials summarising the subject:
- Nilsson Benfatto, M., Öqvist Seimyr, G., Ygge, J., Pansell, T., Rydberg, A., & Jacobson, C. (2016). Screening for dyslexia using eye tracking during reading. PLOS ONE, 11(12), e0165508.
- Ward, L. M., & Kapoula, Z. (2020). Differential diagnosis of vergence and saccade disorders in dyslexia. Scientific Reports, 10(1), 22116.
- Alqahtani, N. D., Alzahrani, B., & Ramzan, M. S. (2023). Deep learning applications for dyslexia prediction. Applied Sciences, 13(5), 2804.
- MacCullagh, L., Bosanquet, A., & Badcock, N. A. (2017). University students with dyslexia: A qualitative exploratory study of learning practices, challenges and strategies. Dyslexia, 23(1), 3–23.
- https://openai.com/form/custom-models
A list of requirements for taking part in the project:
- Intermediate programming skills
and/or
- knowledge of psychology of dyslexia
and/or
- visualization skills
A maximal number of participants: 10
Skills and competences you can learn during the project:
- Gain empathy and awareness of how individuals with dyslexia perceive and interpret text.
- Hands-on experience in training a specialized language model.
- Develop the ability to extract meaningful information from text data, identifying key patterns in dyslexic reading.
- Learn to create impactful visualizations, such as videos or images, to convey dyslexic reading patterns effectively.
- Explore creative solutions for enhancing text comprehension for dyslexic readers, contributing to positive social impact.
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 4: Estimating the amount of recalled information by NLP techniques for free recall psychological research of memory
# THIS PROJECT IS FULLY OCCUPIED
Leader: Michał Domagała1
- Jagiellonian University
Abstract: In research of memory, traditionally there exist two methods of judging memorized content: Explicit paradigms, that task participant in recognising presented stimuli as novel or previously seen or Implicit methods, where subject are asked to fill the collection of object with one from memory. Both methods however bear little resemblance to how humans memorize objects in their daily functioning, as they consist of recognising simple, isolated objects as opposed to complex audiovisual stimuli. Thus, a novel paradigm of recalling from a continuous stream of information, such as movies or audiobooks has been proposed as a more ecological alternative. Here however, the problem of judging the amount of remembered information arises, as it is difficult to use objective measurements as people tend to differ in their ascriptions of importance. We propose to use Natural Language Processing and advanced GPT based language models to develop an index of how much detail have been memorized. In order to achieve this we intend to develop similarity measures between auditory stream and recall that is sensitive to context and works irregardless of the text length. Such a tool would be invaluable in ecological psychological research as it will allow for fast and highly reproducible assessment of the amount of remembered information. To achieve the stated objective, we propose the following implementation plan:
1. Generate the dataset of stories by the virtue of language models such as ChatGPT and ask people for detailed and less detailed summary. Additionally, summaries will be performed by language models.
2. Customly refit GPT model to return a remembrance score – as an estimate of recalled information on the basis of text similarity between original text, GPT made summary, and by referencing detailed and less detailed summaries. Here our work will center on reshaping data and establishing text properties that will be connected to better recall.
3. Compare remembrance score with the judgment of Competent judges and replicate simple experiment of Viewing a standard audiovisual stimuli of “Sherlock” movie and then asking participants to Recall specific scenes in as detailed way as possible.
A list of 1-5 key papers/materials summarising the subject:
- https://openai.com/form/custom-models
- https://www.newscatcherapi.com/blog/ultimate-guide-to-text-similarity-with-python
- Frisoni, M., Di Ghionno, M., Guidotti, R., Tosoni, A., & Sestieri, C. (2023). Effects of a narrative template on memory for the time of movie scenes: Automatic reshaping is independent of consolidation. Psychological Research, 87(2), 598–612.
- Brownlee, K. (2015). The Competent Judge Problem. Ratio, 29(3), 312–326.
A list of requirements for taking part in the project:
- Decent computer skills
- preferable some degree of knowledge about NLP or Psychology
A maximal number of participants: 6
Skills and competences you can learn during the project:
- Basics of NLP processing
- Learning Caveats of memory related psychological reaserch
- GPT prompting and refitting
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 5: Default Mode Network connectivity
# THIS PROJECT IS FULLY OCCUPIED
Leader: Zofia Borzyszkowska1
- Nicolaus Copernicus Univeristy
Abstract: Default Mode Network (DMN) is active during a resting state and is especially helpful in identifying differences between healthy people, and patients with mental diseases. The relationship between resting state and visual input has not been widely researched. In 2022 Yi Wang lead an EEG study aiming to find differences in connectivity in between two states: eyes open and eyes closed [1]. Analysis carried out on the Brainhack Warsaw is a part of an ongoing Neurotech Students Scientific Club project inspired by Wang’s paper. The EEG data for this project has been collected by members of the club on a 32-electrode cap. The goal of this project is to analyze DMN connectivity between two states: eyes closed, eyes opened.
A list of 1-5 key papers/materials summarising the subject:
1. Y. Wang et al., “Open Eyes Increase Neural Oscillation and Enhance Effective Brain Connectivity of the Default Mode Network: Resting-State Electroencephalogram Research,” Front. Neurosci., vol. 16, p. 861247, Apr. 2022, doi: 10.3389/fnins.2022.861247.
A list of requirements for taking part in the project:
- basics of Python would be an advantage
- basics of Matlab would be an advantage
A maximal number of participants: 5
Skills and competences you can learn during the project:
- better understanding of EEG analysis
- basics in Python libraries for connectivity analysis
- basic github
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 6: LoRACraft: does composing diffusion model LoRAs actually work?
Leader: Paweł Pierzchlewicz1
- University of Tübingen and University of Göttingen
Abstract: In the realm of high-quality image generation, diffusion models have demonstrated prowess, notably with the infusion of Low-Rank Adaptation (LoRA). This technique excels at injecting novel concepts into diffusion models, enhancing versatility and creativity. Despite its success in fine-tuning for specific tasks, the potential of composing LoRAs for multi-task capabilities remains underexplored. Enter LoRACraft: a project delving into the uncharted territory of composing LoRAs. Our mission is to scrutinize the limitations, crafting different models to evaluate their individual task performance and their prowess in combination. Drawing a link between LoRAs’ composability and the energy-based perspective on diffusion models, we aim to establish a robust theory explaining the efficacy of combining these models. Our hypothesis posits that composing LoRAs is akin to composing energies, allowing for the flexible combination of LoRAs and achieving superior performance in compositional tasks. LoRACraft seeks to unravel the potential synergy, providing insights into the underexplored realm of LoRA composition for enhanced performance in diffusion models.
A list of 1-5 key papers/materials summarising the subject:
- https://showlab.github.io/Mix-of-Show/
- https://arxiv.org/abs/2307.13269
- https://openreview.net/pdf?id=uWvKBCYh4S
- https://arxiv.org/abs/2106.09685
- https://huggingface.co/docs/diffusers/tutorials/using_peft_for_inference
A list of requirements for taking part in the project:
- Communicative English
- Experience with PyTorch
- Prior experience with diffusion models and LoRAs is a plus
A maximal number of participants: 6
Skills and competences you can learn during the project:
- Learn some theory behind representation learning,
- pratice implementing down-stream tasks that use latent space representations,
- create together new tools for combining latent space representations
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 7: Rhythms of thought: Unraveling human behavior with bayesian models and circadian rhythmicity
Leader: Patrycja Ściślewska1
- Department of Animal Physiology, Faculty of Biology, University of Warsaw; Laboratory of Emotions Neurobiology, Nencki Institute of Experimental Biology PAS
Abstract: Bayesian models are increasingly used across a broad spectrum of the cognitive sciences. This approach aligns with the current trends in data-driven attempts to understand human behavior (Wilson & Collins, 2019). The aim of the project is to develop a mathematical model to describe human behavior (e.g., reaction time in particular conditions, decision-making processes, learning rate) and to determine the relationship between these aspects of human behavior and the features of the biological clock, sleep quality, and personality traits. Are the evening people more willing to take risks? Do morning larks learn faster? How does the negative emotionality affect our reaction time? At the Hackathon, we will use experimental data from state-of-the-art neuroscience tasks (such as the Iowa Gambling Task or the Monetary Incentive Delay Task), which measure individual sensitivity to reward and punishment, the tendency to take risks, or to learn from mistakes. We will choose one of the computational models described in the literature (e.g. Rescorla-Wagner Model (Rescorla & Wagner, 1972), Outcome-Representation Learning Model (Haines et al., 2018)) and modify it to best fit the behavioral data. Then, we will perform computer simulations to validate the model. The results will provide more insight into the role of the circadian rhythmicity and sleep in the proper functioning of cognitive processes in the brains of young people.
A list of 1-5 key papers/materials summarising the subject:
- Robert C Wilson, Anne GE Collins (2019). Ten simple rules for the computational modeling of behavioral data
- L. Griffiths, Thomas; Kemp, Charles; B. Tenenbaum, Joshua (2018). Bayesian models of cognition. Carnegie Mellon University. Journal contribution.
- Haines N, Vassileva J, Ahn WY (2018). The Outcome-Representation Learning Model: A Novel Reinforcement Learning Model of the Iowa Gambling Task. Cogn Sci. doi:10.1111/cogs.12688
- Fabian A. Soto, Edgar H. Vogel, Yerco E. Uribe-Bahamonde, Omar D. Perez (2023). Why is the Rescorla-Wagner model so influential?
A list of requirements for taking part in the project:
- No fear of math will be appreciated 🙂
A maximal number of participants: 10
Skills and competences you can learn during the project:
Participants will learn the most commonly used tasks for measuring human cognitive processes and behavior. Participants will have the opportunity to delve into the world of computational modeling, specifically with Bayesian approaches. They will learn how to choose and adapt existing computational behavior models to fit unique behavioral datasets, fostering a practical understanding of applying theoretical concepts to real-world scenarios. Finally, participants will explore the relationship between circadian rhythmicity, sleep quality, and personality traits in the context of cognitive processes.
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 8: Can AI diagnose mental conditions within a single conversation?
# THIS PROJECT IS FULLY OCCUPIED
Leader: Piotr Migdał PhD1, Michał Jaskólski1
Abstract:Modern large language models (LLMs) exhibit immense prowess in interpreting text data, particularly in natural language. They adeptly extract information, not only from explicit statements but also by reading between the lines, utilizing elements from word usage and syntax to overall coherence. For this exploration, we will utilize GPT-4 by OpenAI, the most potent LLM available to date. It has been trained on extensive datasets, including psychological and therapeutic textbooks, articles, and innumerable real conversations. This project investigates whether this generalist model can effectively assess an individual’s psychological traits. Our aim is to predict various psychological traits, such as the Big Five and Dark Triad, along with Attachment Style, as well as neurodivergences like autism and ADHD. We remain open to predicting other traits and conditions, contingent on the available data and participant interest. Traditional questionnaires use a rigid structure of questions and answers. With the latest AI model, we can transcend this limitation through automatic text analysis and AI-enabled conversations, mimicking a mental care professional’s approach.
- What methods are more effective: text analysis or interactive discussion?
- Which psychological traits and conditions are more accurately predictable?
- How can we optimize prompts for GPT-4 to ensure high-quality responses?
- What is the minimal data required for accurate predictions? – How can we benchmark and validate our findings?
In 2013, simpler machine learning models could predict personality from as few as 10 Facebook likes, more accurately than a work colleague, and with 300 likes, better than a spouse. We seek to discover the extent of advancements possible with far more sophisticated AI and richer data.
A list of 1-5 key papers/materials summarising the subject:
- State of GPT – Andrej Karpathy
- The End of Human Doctors – Understanding Medicine
- Private traits and attributes are predictable from digital records of human behavior
- Towards Conversational Diagnostic AI
- Can Generalist Foundation Models Outcompete Special-Purpose Tuning? Case Study in Medicine
A list of requirements for taking part in the project:
- Communicative English
A maximal number of participants: 10
Skills and competences you can learn during the project:
- Core concepts behind the transformer neural networks, their applications, strengths and weaknesses.
- Programmatically call GPT-4 (more general and powerful than via ChatGPT interference).
- Generating structured data (instead of plain text) via GPT-4.
- Difference between a zero-shot (i.e. no examples given) and a few-shot inference.
- Fundamental limitations of every (existing or future) using only text interface.
- Ethical & safety considerations: biases, misapplications, benefits vs dangers.
- How much can be derived from our digital footprint?
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 9: Large Language Models vs Human Cerebral Cortex: Similarities, Differences, and their Consequences
Leader: Natalia Bielczyk PhD1
Abstract:
Will Large Language Models (LLMs) take our jobs? This is a delicate and complex subject-matter: AI is designed to automate tasks rather than jobs, while most jobs consist of dozens of tasks.
Similarly to other reinforcement learning models, Large Language Models were originally inspired by the human brain. Indeed, human cortical networks and large language models (LLMs) such as GPT share some similarities in their structure, function, and learning mechanisms.
- Structural Similarities: Both human cortical networks and LLMs are composed of interconnected nodes (neurons in the brain, artificial neurons in LLMs) that process and transmit information. Transformer-based LLMs, in particular, have been found to have structurally similar representations to neural response measurements from brain imaging.
- Functional Similarities: Both human brains and LLMs process language in a predictive manner, predicting the next word based on the context. Studies have shown that the activations of LLMs map accurately to areas distributed over the two hemispheres of the brain, suggesting a functional similarity.
- Learning Similarities: Both human brains and LLMs learn from experience. For LLMs, this experience comes in the form of large amounts of text data that they are trained on.
However, there are also profound differences, especially in terms of consciousness, learning mechanisms, and the ability to handle abstract logic and long-term consistency.
- Structural Differences: The human brain is a highly complex, three-dimensional network of neurons with both local and long-range connections, while LLMs are typically organized in layers with connections primarily within and between adjacent layers. Moreover, the brain’s structure is influenced by physical locality and developmental processes, leading to a modular structure.
- Functional Differences: While LLMs excel at tasks like translation and text completion, they struggle with tasks that require abstract logic or long-term consistency, which are areas where the human brain excels. Additionally, LLMs lack the bidirectional connectivity that is believed to be essential for consciousness in the human brain.
- Learning Differences: The human brain learns from a variety of sensory inputs and experiences over a lifetime, while LLMs are trained on specific datasets and their learning is confined to the patterns present in these datasets. Furthermore, the brain is capable of lifelong learning and adaptation, while LLMs’ parameters remain fixed after training. On the other hand, learning in LLMs is fast and widespread as back-propagation mechanism allows for global and fast learning in the network while Hebbian learning in cortical networks is local and slow.
In this project, we will go through the following steps: … Read the rest of abstract here!
A list of 1-5 key papers/materials summarising the subject:
- Vaswani et al. (2017). Attention Is All You Need.
- What is a Large Language Model (LLM).
- Exploring LLMs: Classification, Use Cases, and Challenges.
- Van Essen et al. (1998). Functional and structural mapping of human cerebral cortex: Solutions are in the surfaces.
- S. Schipp. Structure and function of the cerebral cortex.
A list of requirements for taking part in the project:
- Education level: BS / MS in STEM sciences (mathematics, computer science, bioinformatics, neuroscience, cognitive science, etc.)
- strong command of English
- basic programming skills in Python
- basic statistics
A maximal number of participants: 20
Skills and competences you can learn during the project:
The participants will have the opportunity to learn about the cutting-edge standards in Large Language Models: their architecture, complexity, and useful features. At the same time, they can get familiar with the current state of knowledge about the structure and function of the human cerebral cortex, and the state-of-the-art methods for modeling the dynamics of brain networks.
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 10:
Stroke lesion segmentation via unsupervised anomaly detection
Leader: Cemal Koba PhD1
- Sano Center
Abstract: Detection of areas affected by stroke is a crucial aspect for its recovery. Automatization of this task has been a popular challenge, but the ground truth for confirming the performance of the automatic processes was the lesion masks that are manually drawn by human experts. However, stroke lesions may not be always visible to human eye. In this project, we are aiming at automatic lesion segmentation based an unsupervised model. The model will be trained on the healthy brains and the aim will be anomaly detection (stroke lesion, in our case) through generative adversarial networks.
A list of 1-5 key papers/materials summarising the subject:
- https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03936-1
- https://www.sciencedirect.com/science/article/abs/pii/S0165027019304327
- https://www.sciencedirect.com/science/article/pii/S1746809419302228?casa_token=XxbInRp_5a8AAAAA:ioG9pZvhMS4VRi7m8007d6O7ShZ3w88GbR-VgfzB7lL22KileOdgTH5VZkL5J_CSoTzR3ot6Cf8
A list of requirements for taking part in the project:
- Basic understanding of scripting in python,
- 3D image processing,
- deep learning algorithms are appreciated, but not a must.
A maximal number of participants: 8
Skills and competences you can learn during the project:
The parctice will be mostly on the coding part, so the participants will have experience in dealing with a neuroimaging dataset, running a deep learning algortihm in python
Is there a plan for extending this work to a paper in case the results are promising? Yes
Speakers
Opening lecture: Bridging the gap between biology and AI: dendrites, gradients and the quest for Artificial General Intelligence
Ziemowit Sławński
Abstract: Exploring the complexity of biological neurons could change the way we think about building Artificial Intelligence (AI). Neurons are not just simple message carriers; each is a complex network capable of performing advanced computations. This raises a fascinating question: Could insights from studying the structure and function of dendrites open up a new era of more powerful AI? Intelligence is not just about processing information; it has evolved to facilitate meaningful interaction with the environment. During this talk, we will explore how embodied modelling based on complex biological networks could open the door to Artificial General Intelligence (AGI), and ask whether, surprisingly, Large Language Models (LLM) might be able to mimic everything we ever dreamed for AGI but we were too naive to look elsewhere.
Closing lecture: Mind the market - bridging science and business for neuroimaging technology
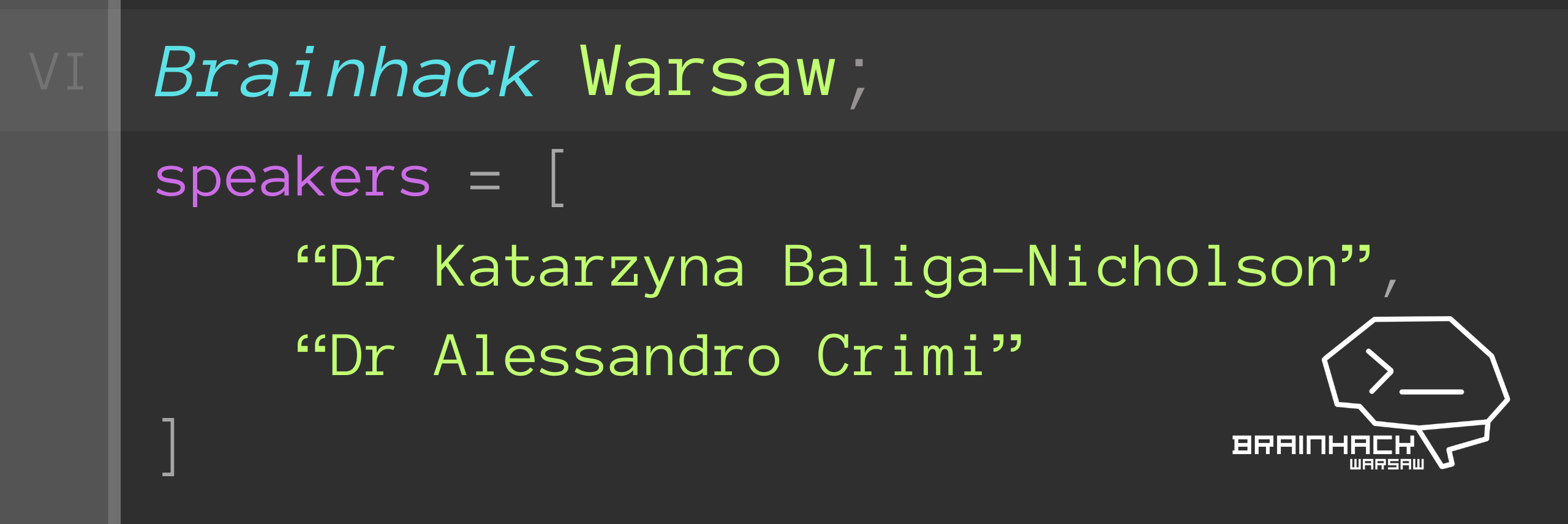
Katarzyna Baliga-Nicholson, PHD
Dr Katarzyna Baliga-Nicholson is a researcher and practitioner with a focus on management and artificial intelligence. She holds a Post Graduate Certificate in Education in Business and Economics from the University of London, a master’s degree in management from Jagiellonian University, and a PhD in digital innovation management, also from Jagiellonian University.Alessandro Crimi, PHD
Dr Alessandro Crimi worked as a postdoctoral researcher in different countries including Italy, France and Switzerland, mostly on clinical neuroimaging projects about multiple sclerosis, Alzheimer’s and Parkinson’s disease and glioma. He holds a PhD in medical imaging from the University of Copenhagen and MBA in international health management from the Swiss Tropical and Public Health Institute (University of Basel).
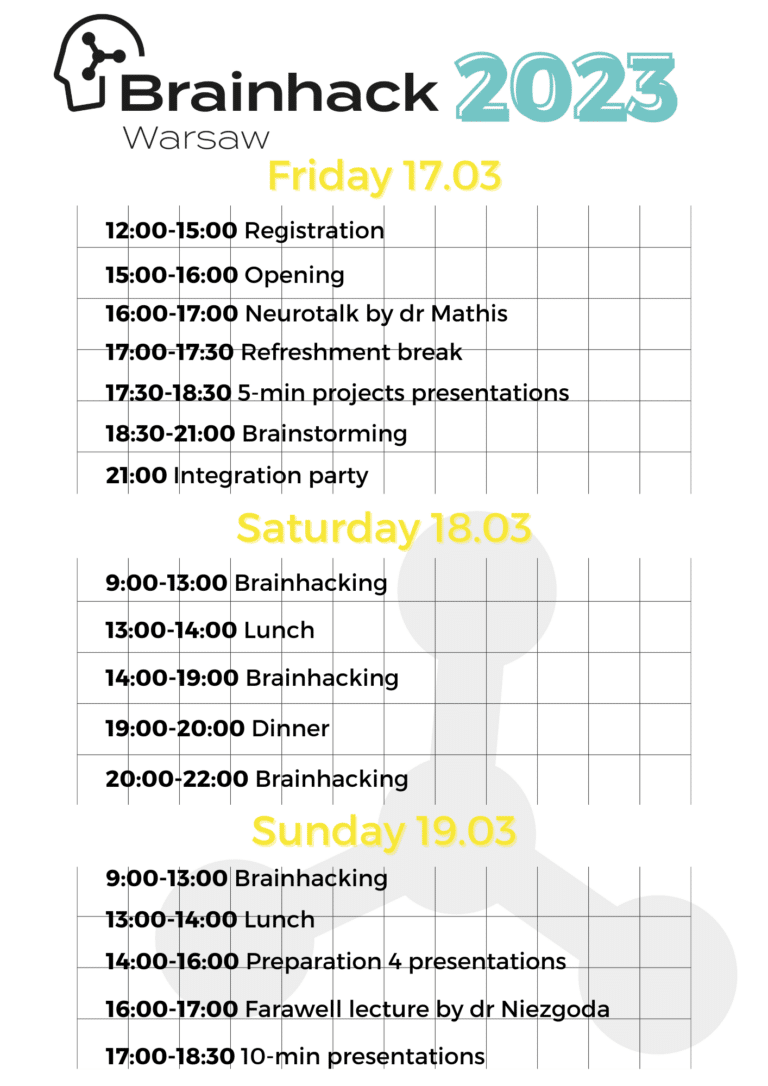
Schedule


Participant Registration
Deadline: 29.02.2024
Registration Form: click here!
Registration take place in two rounds and there is a small registration fee (to cover the catering during the event) for the project participants:
EARLY -> 200 PLN (around 45 EUR) for participants registratered till 4th February
REGULAR -> 225 PLN (around 51 EUR) for participants registered from 5th February to 29th February
Please keep in mind that all of the payments should be made in PLN (polish złoty).
Partners & sponsors
Committee

Kamila Trafna
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

JULIA Jakubowska
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

ALEKSANDRA BARTNIK
Neuroinformatics,
Faculty of Physics, Cognitive Science, Faculty of Psychology, University of Warsaw,
Warsaw, Poland

PAULA BANACH
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Karolina Piwko
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

KATARZYNA WACH
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Maria Waligórska
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Igor Kołodziej
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Gabriela Pawlak
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Julia Pelczar
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Karolina Tymicka
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Krzysztof Wróblewski
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Kornelia Bordzoł
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland
Aleksander Rogowski
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Sylwia Adamus
Neuroinformatics,
Medical University of Warsaw, Faculty of Physics, University of Warsaw,
Warsaw, Poland

Weronika Sarna
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Łukasz Niedźwiedzki
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland
Martyna Trela
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland

Taisiia Prosvirova
Cognitive Science,
Faculty of Psychology, University of Warsaw,
Warsaw, Poland

Zofia Mizgalewicz
Neuroinformatics,
Faculty of Physics, University of Warsaw,
Warsaw, Poland
Advisory board:

Jarosław Żygierewicz, PhD
Neuroinformatics,
Biomedical Physics Division,
Faculty of Physics, University of Warsaw,
Warsaw, Poland
jacek Cyranka, PhD
Assistant professor in Computer Science,
Institute of Informatics,
Faculty of Mathematics, Informatics, and Mechanics, University of Warsaw,
Warsaw, Poland
Venue
University of Warsaw, Faculty of Physics, Pasteura 5, 02-093 Warsaw, Poland.